- Record: found
- Abstract: found
- Article: found
Discriminant canonical tool for inferring the effect of αS1, αS2, β, and κ casein haplotypes and haplogroups on zoometric/linear appraisal breeding values in Murciano-Granadina goats
Read this article at
Abstract
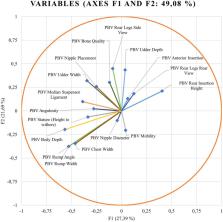
Genomic tools have shown promising results in maximizing breeding outcomes, but their impact has not yet been explored. This study aimed to outline the effect of the individual haplotypes of each component of the casein complex (αS1, β, αS2, and κ-casein) on zoometric/linear appraisal breeding values. A discriminant canonical analysis was performed to study the relationship between the predicted breeding value for 17 zoometric/linear appraisal traits and the aforementioned casein gene haplotypic sequences. The analysis considered a total of 41,323 zoometric/linear appraisal records from 22,727 primiparous does, 17,111 multiparous does, and 1,485 bucks registered in the Murciano-Grandina goat breed herdbook. Results suggest that, although a lack of significant differences ( p > 0.05) was reported across the predictive breeding values of zoometric/linear appraisal traits for αS1, αS2, and κ casein, significant differences were found for β casein ( p < 0.05). The presence of β casein haplotypic sequences GAGACCCC, GGAACCCC, GGAACCTC, GGAATCTC, GGGACCCC, GGGATCTC, and GGGGCCCC, linked to differential combinations of increased quantities of higher quality milk in terms of its composition, may also be connected to increased zoometric/linear appraisal predicted breeding values. Selection must be performed carefully, given the fact that the consideration of apparently desirable animals that present the haplotypic sequence GGGATCCC in the β casein gene, due to their positive predicted breeding values for certain zoometric/linear appraisal traits such as rear insertion height, bone quality, anterior insertion, udder depth, rear legs side view, and rear legs rear view, may lead to an indirect selection against the other zoometric/linear appraisal traits and in turn lead to an inefficient selection toward an optimal dairy morphological type in Murciano-Granadina goats. Contrastingly, the consideration of animals presenting the GGAACCCC haplotypic sequence involves also considering animals that increase the genetic potential for all zoometric/linear appraisal traits, thus making them recommendable as breeding animals. The relevance of this study relies on the fact that the information derived from these analyses will enhance the selection of breeding individuals, in which a desirable dairy type is indirectly sought, through the haplotypic sequences in the β casein locus, which is not currently routinely considered in the Murciano-Granadina goat breeding program.
Related collections
Most cited references64
- Record: found
- Abstract: found
- Article: not found
PLINK: a tool set for whole-genome association and population-based linkage analyses.
- Record: found
- Abstract: not found
- Book: not found
An Introduction to Statistical Learning
- Record: found
- Abstract: not found
- Article: not found