- Record: found
- Abstract: found
- Article: not found
Ivermectin as a potential drug for treatment of COVID-19: an in-sync review with clinical and computational attributes
Read this article at
Abstract
Introduction
COVID-19 cases are on surge; however, there is no efficient treatment or vaccine that can be used for its management. Numerous clinical trials are being reviewed for use of different drugs, biologics, and vaccines in COVID-19. A much empirical approach will be to repurpose existing drugs for which pharmacokinetic and safety data are available, because this will facilitate the process of drug development. The article discusses the evidence available for the use of Ivermectin, an anti-parasitic drug with antiviral properties, in COVID-19.
Methods
A rational review of the drugs was carried out utilizing their clinically significant attributes. A more thorough understanding was met by virtual embodiment of the drug structure and realizable viral targets using artificial intelligence (AI)-based and molecular dynamics (MD)-simulation-based study.
Conclusion
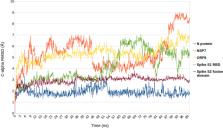
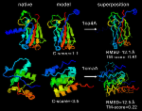
Certain studies have highlighted the significance of ivermectin in COVID-19; however, it requires evidences from more Randomised Controlled Trials (RCTs) and dose- response studies to support its use. In silico-based analysis of ivermectin’s molecular interaction specificity using AI and classical mechanics simulation-based methods indicates positive interaction of ivermectin with viral protein targets, which is leading for SARS-CoV 2 N-protein NTD (nucleocapsid protein N-terminal domain).
Related collections
Most cited references35
- Record: found
- Abstract: found
- Article: not found
The FDA-approved Drug Ivermectin inhibits the replication of SARS-CoV-2 in vitro
- Record: found
- Abstract: found
- Article: found
I-TASSER server for protein 3D structure prediction

- Record: found
- Abstract: found
- Article: found
