- Record: found
- Abstract: found
- Article: found
Cytogenetic study of heptapterids (Teleostei, Siluriformes) with particular respect to the Nemuroglanis subclade
Abstract
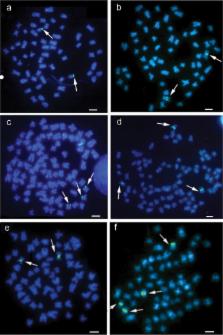
The catfish family Heptapteridae (order Siluriformes ) is endemic to the Neotropics and is one of the most common of the fish families in small bodies of water. Although over 200 species have been identified in this family, very few have been characterized cytogenetically. Here, we analyze the chromosome genomes of four species of Heptapteridae : Cetopsorhamdia iheringi (Schubart & Gomes, 1959), 2n = 58, comprising 28 metacentric (m) + 26 submetacentric (sm) + 4 subtelomeric (st) chromosomes; Pimelodella vittata (Lütken, 1874), 2n = 46, comprising 16m + 22sm + 8st; Rhamdia prope quelen (Quoy & Gaimard, 1824), 2n = 58 comprising 26m + 16sm + 14st + 2 acrocentric; and Rhamdiopsis prope microcephala (Lütken, 1874), 2n = 56, comprising 12m + 30sm + 14st. The nucleolus organizer regions (NORs) were located in a single chromosome pair in all species. The two species that belonged to the subclade Nemuroglanis , Cetopsorhamdia iheringi and Rhamdia prope quelen , had a diploid chromosome number of 58 and an interstitial NOR adjacent to a C + block located on one of the larger chromosome pairs in the complement. Our results from conventional cytogenetic techniques in combination with FISH using 18S and 5S rDNA probes corroborated the taxonomical hypothesis for the formation of the Nemuroglanis subclade.
Related collections
Most cited references34

- Record: found
- Abstract: found
- Article: found
The 5S rDNA family evolves through concerted and birth-and-death evolution in fish genomes: an example from freshwater stingrays
- Record: found
- Abstract: found
- Article: not found
Organization of 5S rDNA in species of the fish Leporinus: two different genomic locations are characterized by distinct nontranscribed spacers.
- Record: found
- Abstract: found
- Article: not found