- Record: found
- Abstract: found
- Article: found
Comparative Analysis Highlights Variable Genome Content of Wheat Rusts and Divergence of the Mating Loci
Abstract
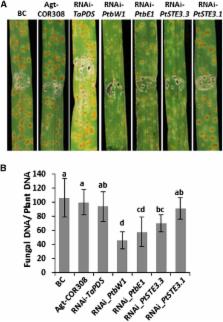
Three members of the Puccinia genus, Puccinia triticina ( Pt), P. striiformis f.sp. tritici ( Pst), and P. graminis f.sp. tritici ( Pgt), cause the most common and often most significant foliar diseases of wheat. While similar in biology and life cycle, each species is uniquely adapted and specialized. The genomes of Pt and Pst were sequenced and compared to that of Pgt to identify common and distinguishing gene content, to determine gene variation among wheat rust pathogens, other rust fungi, and basidiomycetes, and to identify genes of significance for infection. Pt had the largest genome of the three, estimated at 135 Mb with expansion due to mobile elements and repeats encompassing 50.9% of contig bases; in comparison, repeats occupy 31.5% for Pst and 36.5% for Pgt. We find all three genomes are highly heterozygous, with Pst [5.97 single nucleotide polymorphisms (SNPs)/kb] nearly twice the level detected in Pt (2.57 SNPs/kb) and that previously reported for Pgt. Of 1358 predicted effectors in Pt, 784 were found expressed across diverse life cycle stages including the sexual stage. Comparison to related fungi highlighted the expansion of gene families involved in transcriptional regulation and nucleotide binding, protein modification, and carbohydrate degradation enzymes. Two allelic homeodomain pairs, HD1 and HD2, were identified in each dikaryotic Puccinia species along with three pheromone receptor ( STE3) mating-type genes, two of which are likely representing allelic specificities. The HD proteins were active in a heterologous Ustilago maydis mating assay and host-induced gene silencing (HIGS) of the HD and STE3 alleles reduced wheat host infection.
Most cited references52
- Record: found
- Abstract: found
- Article: not found
Comprehensive comparative analysis of strand-specific RNA sequencing methods
- Record: found
- Abstract: found
- Article: not found
DAGchainer: a tool for mining segmental genome duplications and synteny.
- Record: found
- Abstract: found
- Article: not found