- Record: found
- Abstract: found
- Article: found
Combinations of Independent Dominant Loci Conferring Clubroot Resistance in All Four Turnip Accessions ( Brassica rapa) From the European Clubroot Differential Set
Read this article at
Abstract
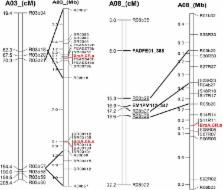
Clubroot disease is devastating to Brassica crop production when susceptible cultivars are planted in infected fields. European turnips are the most resistant sources and their resistance genes have been introduced into other crops such oilseed rape ( Brassica napus L.), Chinese cabbage and other Brassica vegetables. The European clubroot differential (ECD) set contains four turnip accessions (ECD1–4). These ECD turnips exhibited high levels of resistance to clubroot when they were tested under controlled environmental conditions with Canadian field isolates. Gene mapping of the clubroot resistance genes in ECD1–4 were performed and three independent dominant resistance loci were identified. Two resistance loci were mapped on chromosome A03 and the third on chromosome A08. Each ECD turnip accession contained two of these three resistance loci. Some resistance loci were homozygous in ECD accessions while others showed heterozygosity based on the segregation of clubroot resistance in 20 BC 1 families derived from ECD1 to 4. Molecular markers were developed linked to each clubroot resistance loci for the resistance gene introgression in different germplasm.
Related collections
Most cited references24
- Record: found
- Abstract: not found
- Article: not found
The Occurrence and Economic Impact of Plasmodiophora brassicae and Clubroot Disease
- Record: found
- Abstract: found
- Article: not found
Simple sequence repeat-based comparative genomics between Brassica rapa and Arabidopsis thaliana: the genetic origin of clubroot resistance.
- Record: found
- Abstract: not found
- Article: not found