- Record: found
- Abstract: found
- Article: found
Restricted Genetic Variation in Populations of Achatina ( Lissachatina) fulica outside of East Africa and the Indian Ocean Islands Points to the Indian Ocean Islands as the Earliest Known Common Source
Read this article at
Abstract
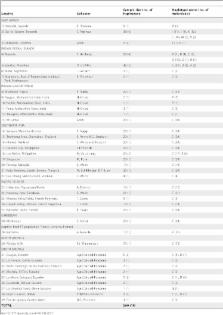
The Giant African Land Snail, Achatina ( = Lissachatina) fulica Bowdich, 1822, is a tropical crop pest species with a widespread distribution across East Africa, the Indian subcontinent, Southeast Asia, the Pacific, the Caribbean, and North and South America. Its current distribution is attributed primarily to the introduction of the snail to new areas by Man within the last 200 years. This study determined the extent of genetic diversity in global A. fulica populations using the mitochondrial 16S ribosomal RNA gene. A total of 560 individuals were evaluated from 39 global populations obtained from 26 territories. Results reveal 18 distinct A. fulica haplotypes; 14 are found in East Africa and the Indian Ocean islands, but only two haplotypes from the Indian Ocean islands emerged from this region, the C haplotype, now distributed across the tropics, and the D haplotype in Ecuador and Bolivia. Haplotype E from the Philippines, F from New Caledonia and Barbados, O from India and Q from Ecuador are variants of the emergent C haplotype. For the non-native populations, the lack of genetic variation points to founder effects due to the lack of multiple introductions from the native range. Our current data could only point with certainty to the Indian Ocean islands as the earliest known common source of A. fulica across the globe, which necessitates further sampling in East Africa to determine the source populations of the emergent haplotypes.
Related collections
Most cited references10
- Record: found
- Abstract: not found
- Article: not found
The Staden package, 1998.
- Record: found
- Abstract: not found
- Article: not found
A simple method of preparing plant samples for PCR.
- Record: found
- Abstract: found
- Article: not found