- Record: found
- Abstract: found
- Article: found
Quantitation of the cellular content of saliva and buccal swab samples
Read this article at
Abstract
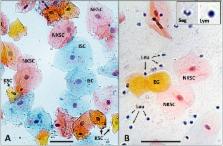
Buccal swabs and saliva are the two most common oral sampling methods used for medical research. Often, these samples are used interchangeably, despite previous evidence that both contain buccal cells and blood leukocytes in different proportions. For some research, such as epigenetic studies, the cell types contributing to the analysis are highly relevant. We collected such samples from twelve children and twenty adults and, using Papanicolaou staining, measured the proportions of epithelial cells and leukocytes through microscopy. To our knowledge, no studies have compared cellular heterogeneity in buccal swab and saliva samples from adults and children. We confirmed that buccal swabs contained a higher proportion of epithelial cells than saliva and that children have a greater proportion of such cells in saliva compared to adults. At this level of resolution, buccal swabs and saliva contained similar epithelial cell subtypes. Gingivitis in children was associated with a higher proportion of leukocytes in saliva samples but not in buccal swabs. Compared to more detailed and costly methods such as flow cytometry or deconvolution methods used in epigenomic analysis, the procedure described here can serve as a simple and low-cost method to characterize buccal and saliva samples. Microscopy provides a low-cost tool to alert researchers to the presence of oral inflammation which may affect a subset of their samples. This knowledge might be highly relevant to their specific research questions, may assist with sample selection and thus might be crucial information despite the ability of data deconvolution methods to correct for cellular heterogeneity.
Related collections
Most cited references22
- Record: found
- Abstract: found
- Article: not found
DNA extracted from saliva for methylation studies of psychiatric traits: evidence tissue specificity and relatedness to brain.

- Record: found
- Abstract: found
- Article: found