- Record: found
- Abstract: found
- Article: found
A global checklist of the Bombycoidea (Insecta: Lepidoptera)
Abstract
Background
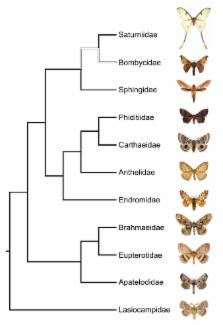
Bombycoidea is an ecologically diverse and speciose superfamily of Lepidoptera . The superfamily includes many model organisms, but the taxonomy and classification of the superfamily has remained largely in disarray. Here we present a global checklist of Bombycoidea . Following Zwick (2008) and Zwick et al. (2011), ten families are recognized: Anthelidae , Apatelodidae , Bombycidae , Brahmaeidae , Carthaeidae , Endromidae , Eupterotidae , Phiditiidae , Saturniidae and Sphingidae . The former families Lemoniidae and Mirinidae are included within Brahmaeidae and Endromidae respectively. The former bombycid subfamilies Oberthueriinae and Prismostictinae are also treated as synonyms of Endromidae , and the former bombycine subfamilies Apatelodinae and Phitditiinae are treated as families.
New information
This checklist represents the first effort to synthesize the current taxonomic treatment of the entire superfamily. It includes 12,159 names and references to their authors, and it accounts for the recent burst in species and subspecies descriptions within family Saturniidae (ca. 1,500 within the past 10 years) and to a lesser extent in Sphingidae (ca. 250 species over the same period). The changes to the higher classification of Saturniidae proposed by Nässig et al. (2015) are rejected as premature and unnecessary. The new tribes, subtribes and genera described by Cooper (2002) are here treated as junior synonyms. We also present a new higher classification of Sphingidae , based on Kawahara et al. (2009), Barber and Kawahara (2013) and a more recent phylogenomic study by Breinholt et al. (2017), as well as a reviewed genus and species level classification, as documented by Kitching (2018).
Related collections
Most cited references35

- Record: found
- Abstract: found
- Article: found
A Large-Scale, Higher-Level, Molecular Phylogenetic Study of the Insect Order Lepidoptera (Moths and Butterflies)
- Record: found
- Abstract: found
- Article: not found
Order Lepidoptera Linnaeus, 1758. In: Zhang, Z.-Q. (Ed.) Animal biodiversity: An outline of higher-level classification and survey of taxonomic richness
- Record: found
- Abstract: found
- Article: not found