- Record: found
- Abstract: found
- Article: found
A multi-tissue multi-omics analysis reveals distinct kineztics in entrainment of diurnal transcriptomes by inverted feeding

Read this article at
Summary
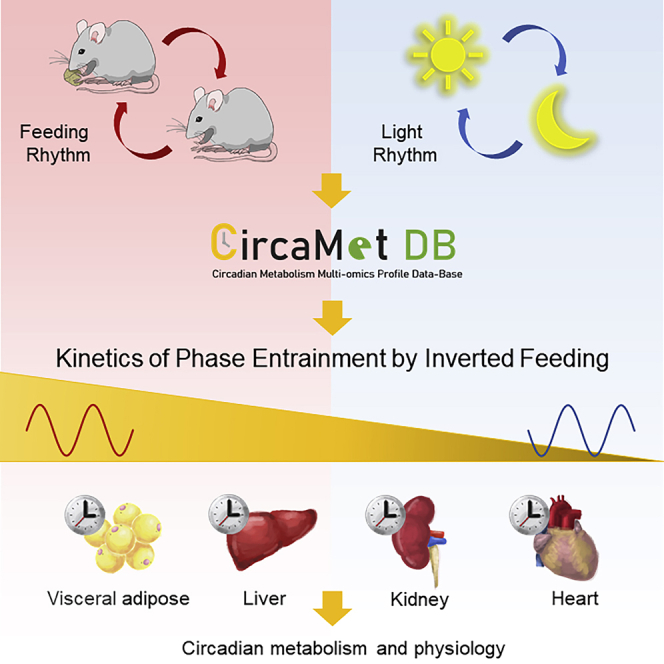
Time of eating synchronizes circadian rhythms of metabolism and physiology. Inverted feeding can uncouple peripheral circadian clocks from the central clock located in the suprachiasmatic nucleus. However, system-wide changes of circadian metabolism and physiology entrained to inverted feeding in peripheral tissues remain largely unexplored. Here, we performed a 24-h global profiling of transcripts and metabolites in mouse peripheral tissues to study the transition kinetics during inverted feeding, and revealed distinct kinetics in phase entrainment of diurnal transcriptomes by inverted feeding, which graded from fat tissue (near-completely entrained), liver, kidney, to heart. Phase kinetics of tissue clocks tracked with those of transcriptomes and were gated by light-related cues. Integrated analysis of transcripts and metabolites demonstrated that fatty acid oxidation entrained completely to inverted feeding in heart despite the slow kinetics/resistance of the heart clock to entrainment by feeding. This multi-omics resource defines circadian signatures of inverted feeding in peripheral tissues ( www.CircaMetDB.org.cn).
Graphical abstract
Highlights
-
•
A multi-omics analysis of food entrainment in mouse peripheral tissues
-
•
Inverted feeding rhythm entrains diurnal transcriptomes with distinct kinetics
-
•
Phase kinetics of tissue clocks is conditioned by constant light
-
•
Cardiac metabolism entrains to feeding fast with slow kinetics of the heart clock
Abstract
Animal Physiology ; Systems Biology ; Metabolomics ; Transcriptomics
Related collections
Most cited references55
- Record: found
- Abstract: found
- Article: not found
Transcriptional architecture of the mammalian circadian clock
- Record: found
- Abstract: found
- Article: not found
A circadian gene expression atlas in mammals: implications for biology and medicine.
- Record: found
- Abstract: found
- Article: not found