- Record: found
- Abstract: found
- Article: found
Rapid Multiplex Small DNA Sequencing on the MinION Nanopore Sequencing Platform
Abstract
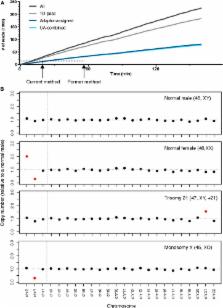
Real-time sequencing of short DNA reads has a wide variety of clinical and research applications including screening for mutations, target sequences and aneuploidy. We recently demonstrated that MinION, a nanopore-based DNA sequencing device the size of a USB drive, could be used for short-read DNA sequencing. In this study, an ultra-rapid multiplex library preparation and sequencing method for the MinION is presented and applied to accurately test normal diploid and aneuploidy samples’ genomic DNA in under three hours, including library preparation and sequencing. This novel method shows great promise as a clinical diagnostic test for applications requiring rapid short-read DNA sequencing.
Most cited references13
- Record: found
- Abstract: found
- Article: not found
Improved data analysis for the MinION nanopore sequencer
- Record: found
- Abstract: found
- Article: not found
Solid-State and Biological Nanopore for Real-Time Sensing of Single Chemical and Sequencing of DNA.
- Record: found
- Abstract: not found
- Article: not found