- Record: found
- Abstract: found
- Article: found
A simple surface plasmon resonance biosensor for detection of PML/RARα based on heterogeneous fusion gene-triggered nonlinear hybridization chain reaction
Read this article at
Abstract
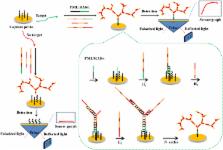
In this work, a simple and enzyme-free surface plasmon resonance (SPR) biosensing strategy has been developed for highly sensitive detection of two major PML/RARα (promyelocytic leukemia, retinoic acid receptor alpha) subtypes based on the heterogeneous fusion gene-triggered nonlinear hybridization chain reaction (HCR). On the gold chip surface, the cascade self-assembly process is triggered after the introduction of PML/RARα. The different fragments of PML/RARα can specifically hybridize with capture probes (Cp) immobilized on the chip and the hybridization DNA 1 (H 1). Then, the nonlinear HCR is initiated by the complex of Cp-PML/RARα-H 1 with the introduction of two hybridization DNA chains (H 2 and H 3). As a result, a dendritic nanostructure is achieved on the surface of chip, leading to a significant SPR amplification signal owing to its high molecular weight. The developed method shows good specificity and high sensitivity with detection limit of 0.72 pM for “L” subtype and 0.65 pM for “S” subtype. Moreover, this method has been successfully applied for efficient identification of clinical positive and negative PCR samples of the PML/RARα subtype. Thus, this developed biosensing strategy presents a potential platform for analysis of fusion gene and early diagnosis of clinical disease.
Related collections
Most cited references36
- Record: found
- Abstract: not found
- Article: not found
Surface plasmon resonance sensors for detection of chemical and biological species.
- Record: found
- Abstract: found
- Article: not found
Nanomaterials. Programmable materials and the nature of the DNA bond.
- Record: found
- Abstract: found
- Article: not found