- Record: found
- Abstract: found
- Article: found
Fungal community analysis by high-throughput sequencing of amplified markers – a user's guide
Read this article at
Abstract
-
Novel high-throughput sequencing methods outperform earlier approaches in terms of resolution and magnitude. They enable identification and relative quantification of community members and offer new insights into fungal community ecology. These methods are currently taking over as the primary tool to assess fungal communities of plant-associated endophytes, pathogens, and mycorrhizal symbionts, as well as free-living saprotrophs.
-
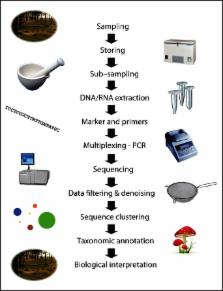
Taking advantage of the collective experience of six research groups, we here review the different stages involved in fungal community analysis, from field sampling via laboratory procedures to bioinformatics and data interpretation. We discuss potential pitfalls, alternatives, and solutions.
-
Highlighted topics are challenges involved in: obtaining representative DNA/RNA samples and replicates that encompass the targeted variation in community composition, selection of marker regions and primers, options for amplification and multiplexing, handling of sequencing errors, and taxonomic identification.
-
Without awareness of methodological biases, limitations of markers, and bioinformatics challenges, large-scale sequencing projects risk yielding artificial results and misleading conclusions.
Related collections
Most cited references90
- Record: found
- Abstract: not found
- Book Chapter: not found
AMPLIFICATION AND DIRECT SEQUENCING OF FUNGAL RIBOSOMAL RNA GENES FOR PHYLOGENETICS
- Record: found
- Abstract: found
- Article: not found
Fast UniFrac: Facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data

- Record: found
- Abstract: found
- Article: found
