- Record: found
- Abstract: found
- Article: found
Personalized whole‐body models integrate metabolism, physiology, and the gut microbiome
Read this article at
Abstract
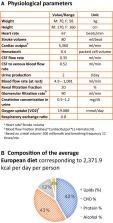
Comprehensive molecular‐level models of human metabolism have been generated on a cellular level. However, models of whole‐body metabolism have not been established as they require new methodological approaches to integrate molecular and physiological data. We developed a new metabolic network reconstruction approach that used organ‐specific information from literature and omics data to generate two sex‐specific whole‐body metabolic ( WBM) reconstructions. These reconstructions capture the metabolism of 26 organs and six blood cell types. Each WBM reconstruction represents whole‐body organ‐resolved metabolism with over 80,000 biochemical reactions in an anatomically and physiologically consistent manner. We parameterized the WBM reconstructions with physiological, dietary, and metabolomic data. The resulting WBM models could recapitulate known inter‐organ metabolic cycles and energy use. We also illustrate that the WBM models can predict known biomarkers of inherited metabolic diseases in different biofluids. Predictions of basal metabolic rates, by WBM models personalized with physiological data, outperformed current phenomenological models. Finally, integrating microbiome data allowed the exploration of host–microbiome co‐metabolism. Overall, the WBM reconstructions, and their derived computational models, represent an important step toward virtual physiological humans.
Abstract
Related collections
Most cited references67
- Record: found
- Abstract: found
- Article: not found
The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon.
- Record: found
- Abstract: found
- Article: not found
A protocol for generating a high-quality genome-scale metabolic reconstruction.
- Record: found
- Abstract: found
- Article: not found
