- Record: found
- Abstract: found
- Article: found
Regulation of cell cycle drivers by Cullin-RING ubiquitin ligases
Read this article at
Abstract
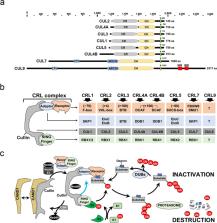
The last decade has revealed new roles for Cullin-RING ubiquitin ligases (CRLs) in a myriad of cellular processes, including cell cycle progression. In addition to CRL1, also named SCF (SKP1-Cullin 1-F box protein), which has been known for decades as an important factor in the regulation of the cell cycle, it is now evident that all eight CRL family members are involved in the intricate cellular pathways driving cell cycle progression. In this review, we summarize the structure of CRLs and their functions in driving the cell cycle. We focus on how CRLs target key proteins for degradation or otherwise alter their functions to control the progression over the various cell cycle phases leading to cell division. We also summarize how CRLs and the anaphase-promoting complex/cyclosome (APC/C) ligase complex closely cooperate to govern efficient cell cycle progression.
Cell biology: Cell cycle mediators offer therapeutic targets
A large group of protein complexes involved in regulating cell division offers an array of promising new drug targets for treating cancer and neurodegeneration. Mirit Aladjem, Sang-Min Jang and colleagues from the US National Cancer Institute in Bethesda, USA, review the structure and function of cullin-RING ubiquitin ligases (CRLs), a “super-family” of multi-subunit enzymes that serve as critical checkpoints on the cell cycle and DNA replication. CRLs promote progression through the cell cycle by targeting other key proteins for degradation or modification. They also work with other protein complexes to ensure genome integrity in the face of DNA damage. Because CRL components are often mutated or otherwise dysregulated in tumors and other diseased cells, small-molecule inhibitors and modulators of CRL activity could have therapeutic benefit for patients.
Related collections
Most cited references124
- Record: found
- Abstract: found
- Article: not found
p21 in cancer: intricate networks and multiple activities.
- Record: found
- Abstract: found
- Article: not found
The myeloma drug lenalidomide promotes the cereblon-dependent destruction of Ikaros proteins.
- Record: found
- Abstract: found
- Article: not found