- Record: found
- Abstract: found
- Article: not found
miR-145 and miR-143 Regulate Smooth Muscle Cell Fate Decisions
Read this article at
SUMMARY
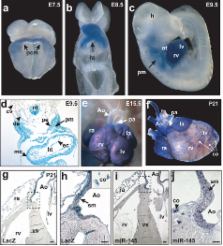
microRNAs are regulators of myriad cellular events, but evidence for a single microRNA that can efficiently differentiate multipotent cells into a specific lineage or regulate direct reprogramming of cells into an alternate cell fate has been elusive. Here, we show that miR-145 and miR-143 are co-transcribed in multipotent cardiac progenitors before becoming localized to smooth muscle cells, including neural crest stem cell–derived vascular smooth muscle cells. miR-145 and miR-143 were direct transcriptional targets of serum response factor, myocardin and Nkx2.5, and were downregulated in injured or atherosclerotic vessels containing proliferating, less differentiated smooth muscle cells. miR-145 was necessary for myocardin-induced reprogramming of adult fibroblasts into smooth muscle cells and sufficient to induce differentiation of multipotent neural crest stem cells into vascular smooth muscle. Furthermore, miR-145 and miR-143 cooperatively targeted a network of transcription factors, including Klf4, myocardin, and Elk-1 to promote differentiation and repress proliferation of smooth muscle cells. These findings demonstrate that miR-145 can direct the smooth muscle fate and that miR-145 and miR-143 function to regulate the quiescent versus proliferative phenotype of smooth muscle cells.
Related collections
Most cited references24
- Record: found
- Abstract: found
- Article: not found
Switching from repression to activation: microRNAs can up-regulate translation.
- Record: found
- Abstract: found
- Article: not found
Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis.
- Record: found
- Abstract: found
- Article: not found
