- Record: found
- Abstract: found
- Article: found
Review of Antimicrobial Resistance in the Environment and Its Relevance to Environmental Regulators
Read this article at
Abstract
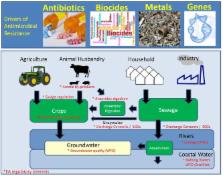
The environment is increasingly being recognized for the role it might play in the global spread of clinically relevant antibiotic resistance. Environmental regulators monitor and control many of the pathways responsible for the release of resistance-driving chemicals into the environment (e.g., antimicrobials, metals, and biocides). Hence, environmental regulators should be contributing significantly to the development of global and national antimicrobial resistance (AMR) action plans. It is argued that the lack of environment-facing mitigation actions included in existing AMR action plans is likely a function of our poor fundamental understanding of many of the key issues. Here, we aim to present the problem with AMR in the environment through the lens of an environmental regulator, using the Environment Agency (England’s regulator) as an example from which parallels can be drawn globally. The issues that are pertinent to environmental regulators are drawn out to answer: What are the drivers and pathways of AMR? How do these relate to the normal work, powers and duties of environmental regulators? What are the knowledge gaps that hinder the delivery of environmental protection from AMR? We offer several thought experiments for how different mitigation strategies might proceed. We conclude that: (1) AMR Action Plans do not tackle all the potentially relevant pathways and drivers of AMR in the environment; and (2) AMR Action Plans are deficient partly because the science to inform policy is lacking and this needs to be addressed.
Related collections
Most cited references202
- Record: found
- Abstract: found
- Article: not found
Comprehensive evaluation of antibiotics emission and fate in the river basins of China: source analysis, multimedia modeling, and linkage to bacterial resistance.
- Record: found
- Abstract: found
- Article: not found
Belowground biodiversity and ecosystem functioning.
- Record: found
- Abstract: found
- Article: not found