- Record: found
- Abstract: found
- Article: found
Inhibitors of Bcl-2 and Bruton’s tyrosine kinase synergize to abrogate diffuse large B-cell lymphoma growth in vitro and in orthotopic xenotransplantation models
Read this article at
Abstract
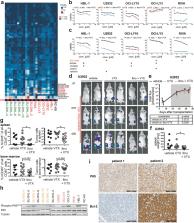
Numerous targeted therapies have been developed for diffuse large B-cell lymphoma, but the results of late-stage clinical trials were mostly disappointing and have led to very few new regulatory approvals. Here, we use single and combinatorial drug response profiling to show that the combined inhibition of the anti-apoptotic protein Bcl-2 and of the tyrosine kinase BTK with the small molecules venetoclax and ibrutinib efficiently kills DLBCL cells in vitro. High Bcl-2 expression due to either BCL2 amplifications or translocations, in conjunction with chronic active BCR signaling accurately predict responses to dual Bcl-2/BTK inhibition. Orthotopic xenotransplantation and patient-derived xenograft models confirm that the combinatorial is superior to single-agent treatment in reducing the lymphoma burden. Combinatorial treatment further efficiently overcomes both primary and acquired resistance to venetoclax, which we could link to reduced expression of the Bcl-2 family members Bcl-X L and Bcl-2A1 under ibrutinib. We found in a Swiss DLBCL cohort that ~15% of patients are projected to respond to the venetoclax/ibrutinib combination based on their high Bcl-2 expression and nuclear NF-κB localization. Our data show that drug sensitivities exposed by drug response profiling can be attributed to specific mutational signatures and immunohistochemical biomarkers, and point to combined Bcl-2/BTK inhibition as a promising therapeutic strategy in DLBCL.
Related collections
Most cited references31
- Record: found
- Abstract: found
- Article: not found
Genetics and Pathogenesis of Diffuse Large B-Cell Lymphoma
- Record: found
- Abstract: found
- Article: not found
Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling.
- Record: found
- Abstract: found
- Article: not found