- Record: found
- Abstract: found
- Article: found
Rectal Swabs for Analysis of the Intestinal Microbiota
Read this article at
Abstract
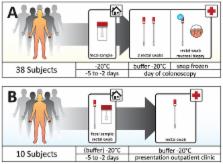
The composition of the gut microbiota is associated with various disease states, most notably inflammatory bowel disease, obesity and malnutrition. This underlines that analysis of intestinal microbiota is potentially an interesting target for clinical diagnostics. Currently, the most commonly used sample types are feces and mucosal biopsy specimens. Because sampling method, storage and processing of samples impact microbiota analysis, each sample type has its own limitations. An ideal sample type for use in routine diagnostics should be easy to obtain in a standardized fashion without perturbation of the microbiota. Rectal swabs may satisfy these criteria, but little is known about microbiota analysis on these sample types. In this study we investigated the characteristics and applicability of rectal swabs for gut microbiota profiling in a clinical routine setting in patients presenting with various gastro-intestinal disorders. We found that rectal swabs appeared to be a convenient means of sampling the human gut microbiota. Swabs can be performed on demand, whenever a patient presents; swab-derived microbiota profiles are reproducible, whether they are gathered at home by patients or by medical professionals in an outpatient setting and may be ideally suited for clinical diagnostics and large-scale studies.
Related collections
Most cited references10
- Record: found
- Abstract: found
- Article: not found
Effect of storage conditions on the assessment of bacterial community structure in soil and human-associated samples.
- Record: found
- Abstract: found
- Article: not found
Freezing fecal samples prior to DNA extraction affects the Firmicutes to Bacteroidetes ratio determined by downstream quantitative PCR analysis.

- Record: found
- Abstract: found
- Article: found