- Record: found
- Abstract: found
- Article: found
Graph Theory Enables Drug Repurposing – How a Mathematical Model Can Drive the Discovery of Hidden Mechanisms of Action
Abstract
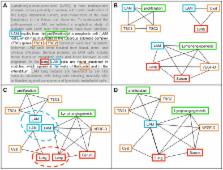
We introduce a methodology to efficiently exploit natural-language expressed biomedical knowledge for repurposing existing drugs towards diseases for which they were not initially intended. Leveraging on developments in Computational Linguistics and Graph Theory, a methodology is defined to build a graph representation of knowledge, which is automatically analysed to discover hidden relations between any drug and any disease: these relations are specific paths among the biomedical entities of the graph, representing possible Modes of Action for any given pharmacological compound. We propose a measure for the likeliness of these paths based on a stochastic process on the graph. This measure depends on the abundance of indirect paths between a peptide and a disease, rather than solely on the strength of the shortest path connecting them. We provide real-world examples, showing how the method successfully retrieves known pathophysiological Mode of Action and finds new ones by meaningfully selecting and aggregating contributions from known bio-molecular interactions. Applications of this methodology are presented, and prove the efficacy of the method for selecting drugs as treatment options for rare diseases.
Related collections
Most cited references44
- Record: found
- Abstract: found
- Article: not found
Structural mechanism for STI-571 inhibition of abelson tyrosine kinase.
- Record: found
- Abstract: found
- Article: not found
Drug-target interaction prediction by random walk on the heterogeneous network.
- Record: found
- Abstract: found
- Article: not found