- Record: found
- Abstract: found
- Article: found
miR-193b regulates tumorigenesis in liposarcoma cells via PDGFR, TGFβ, and Wnt signaling
Read this article at
Abstract
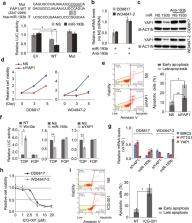
Liposarcoma is the most common soft tissue sarcoma. Molecularly targeted therapeutics have had limited efficacy in liposarcomas, in part because of inadequate knowledge of the complex molecular alterations in these tumors. Our recent study revealed the tumor suppressive function of miR-193b in liposarcoma. Considering the biological and clinical heterogeneity of liposarcoma, here, we confirmed the under-expression of miR-193b in additional patient liposarcoma samples and cell lines. Based on STRING analysis of protein-protein interactions among the reported putative miR-193b targets, we validated three: PDGFRβ, SMAD4, and YAP1, belonging to strongly interacting pathways (focal adhesion, TGFβ, and Hippo, respectively). We show that all three are directly targeted by miR-193b in liposarcoma. Inhibition of PDGFRβ reduces liposarcoma cell viability and increases adipogenesis. Knockdown of SMAD4 promotes adipogenic differentiation. miR-193b targeting of the Hippo signaling effector YAP1 indirectly inhibits Wnt/β-catenin signaling. Both a PDGFR inhibitor (CP-673451) and a Wnt/ β-catenin inhibitor (ICG-001) had potent inhibitory effects on liposarcoma cells, suggesting their potential application in liposarcoma treatment. In summary, we demonstrate that miR-193b controls cell growth and differentiation in liposarcoma by targeting multiple key components (PDGFRβ, SMAD4, and YAP1) in several oncogenic signaling pathways.
Related collections
Most cited references36
- Record: found
- Abstract: found
- Article: not found
FAK integrates growth-factor and integrin signals to promote cell migration.
- Record: found
- Abstract: found
- Article: not found
Sarcoma classification: an update based on the 2013 World Health Organization Classification of Tumors of Soft Tissue and Bone.
- Record: found
- Abstract: found
- Article: not found
