- Record: found
- Abstract: found
- Article: found
Nano-seq analysis reveals different functional tendency between exosomes and microvesicles derived from hUMSC
Read this article at
Abstract
Background
Extracellular vesicles (EVs) from human umbilical cord mesenchymal stem cells (hUMSCs) are widely considered to be the best mediators for cell-free therapy. An understanding of their composition, especially RNA, is particularly important for the safe and precise application of EVs. Up to date, the knowledge of their RNA components is limited to NGS sequencing and cannot provide a comprehensive transcriptomic landscape, especially the long and full-length transcripts. Our study first focused on the transcriptomic profile of hUMSC-EVs based on nanopore sequencing.
Methods
In this study, different EV subtypes (exosomes and microvesicles) derived from hUMSCs were isolated and identified by density gradient centrifugation. Subsequently, the realistic long transcriptomic profile in different subtypes of hUMSC-EVs was systematically compared by nanopore sequencing and bioinformatic analysis.
Results
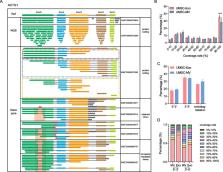
Abundant transcript variants were identified in EVs by nanopore sequencing, 69.34% of which transcripts were fragmented. A series of full-length and long transcripts was also observed and showed a significantly higher proportion of intact or near-complete transcripts in exosomes than that in microvesicles derived from hUMSCs. Although the composition of RNA biotypes transported by different EV subtypes was similar, the distribution of transcripts and genes revealed the inter-heterogeneity and intra-stability between exosomes and microvesicles. Further, 85 different expressed transcripts (56 genes) and 7 fusion genes were identified. Pathway enrichment analysis showed that upregulated-expressed genes in microvesicles were mainly enriched in multiple neurodegenerative diseases, while upregulated-expressed genes in exosomes were mainly enriched in neutrophil extracellular trap formation, suggesting different functional tendencies of EV subtypes.
Conclusions
This study provides a novel understanding of different types of hUMSC-EVs, which not only suggests different transcriptome sorting mechanisms between exosomes and microvesicles, but also shows that different EV subtypes from the same source have different physiological functions, suggesting distinct clinical application prospects.
Related collections
Most cited references38
- Record: found
- Abstract: found
- Article: not found
Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells.

- Record: found
- Abstract: found
- Article: found
Exosome and Exosomal MicroRNA: Trafficking, Sorting, and Function
- Record: found
- Abstract: found
- Article: not found
