- Record: found
- Abstract: found
- Article: not found
Metagenomic Analysis of Fungal Diversity on Strawberry Plants and the Effect of Management Practices on the Fungal Community Structure of Aerial Organs
Read this article at
Abstract
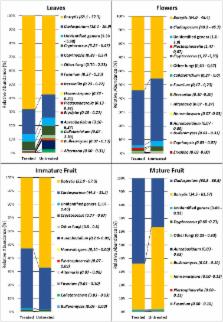
An amplicon metagenomic approach based on the ITS2 region of fungal rDNA was used to identify the composition of fungal communities associated with different strawberry organs (leaves, flowers, immature and mature fruits), grown on a farm using management practices that entailed the routine use of various chemical pesticides. ITS2 sequences clustered into 316 OTUs and Ascomycota was the dominant phyla (95.6%) followed by Basidiomycota (3.9%). Strawberry plants supported a high diversity of microbial organisms, but two genera, Botrytis and Cladosporium, were the most abundant, representing 70–99% of the relative abundance (RA) of all detected sequences. According to alpha and beta diversity analyses, strawberry organs displayed significantly different fungal communities with leaves having the most diverse fungal community, followed by flowers, and fruit. The interruption of chemical treatments for one month resulted in a significant modification in the structure of the fungal community of leaves and flowers while immature and mature fruit were not significantly affected. Several plant pathogens of other plant species, that would not be intuitively expected to be present on strawberry plants such as Erysiphe, were detected, while some common strawberry pathogens, such as Rhizoctonia, were less evident or absent.
Related collections
Most cited references20
- Record: found
- Abstract: found
- Article: not found
Rapid denoising of pyrosequencing amplicon data: exploiting the rank-abundance distribution

- Record: found
- Abstract: found
- Article: found
The Colletotrichum acutatum species complex

- Record: found
- Abstract: found
- Article: found
