- Record: found
- Abstract: found
- Article: not found
Zinc deprivation of methanol fed anaerobic granular sludge bioreactors
Read this article at
Abstract
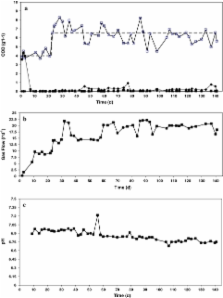
The effect of omitting zinc from the influent of mesophilic (30 °C) methanol fed upflow anaerobic sludge bed (UASB) reactors, and latter zinc supplementation to the influent to counteract the deprivation, was investigated by coupling the UASB reactor performance to the microbial ecology of the bioreactor sludge. Limitation of the specific methanogenic activity (SMA) on methanol due to the absence of zinc from the influent developed after 137 days of operation. At that day, the SMA in medium with a complete trace metal solution except Zn was 3.4 g CH 4-COD g VSS −1 day −1, compared to 4.2 g CH 4-COD g VSS −1 day −1 in a medium with a complete (including zinc) trace metal solution. The methanol removal capacity during these 137 days was 99% and no volatile fatty acids accumulated. Two UASB reactors, inoculated with the zinc-deprived sludge, were operated to study restoration of the zinc limitation by zinc supplementation to the bioreactor influent. In a first reactor, no changes to the operational conditions were made. This resulted in methanol accumulation in the reactor effluent after 12 days of operation, which subsequently induced acetogenic activity 5 days after the methanol accumulation started. Methanogenesis could not be recovered by the continuous addition of 0.5 μM ZnCl 2 to the reactor for 13 days. In the second reactor, 0.5 μM ZnCl 2 was added from its start-up. Although the reactor stayed 10 days longer methanogenically than the reactor operated without zinc, methanol accumulation was observed in this reactor (up to 1.1 g COD-MeOH L −1) as well. This study shows that zinc limitation can induce failure of methanol fed UASB reactors due to acidification, which cannot be restored by resuming the continuous supply of the deprived metal.
Related collections
Most cited references39
- Record: found
- Abstract: found
- Article: not found
Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms.
- Record: found
- Abstract: found
- Article: not found
