- Record: found
- Abstract: found
- Article: not found
Emerging Coxsackievirus A6 Causing Hand, Foot and Mouth Disease, Vietnam
Read this article at
Abstract
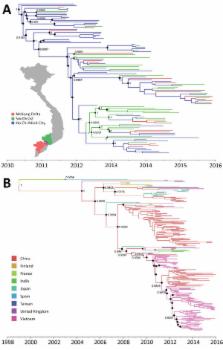
Hand, foot and mouth disease (HFMD) is a major public health issue in Asia and has global pandemic potential. Coxsackievirus A6 (CV-A6) was detected in 514/2,230 (23%) of HFMD patients admitted to 3 major hospitals in southern Vietnam during 2011–2015. Of these patients, 93 (18%) had severe HFMD. Phylogenetic analysis of 98 genome sequences revealed they belonged to cluster A and had been circulating in Vietnam for 2 years before emergence. CV-A6 movement among localities within Vietnam occurred frequently, whereas viral movement across international borders appeared rare. Skyline plots identified fluctuations in the relative genetic diversity of CV-A6 corresponding to large CV-A6–associated HFMD outbreaks worldwide. These data show that CV-A6 is an emerging pathogen and emphasize the necessity of active surveillance and understanding the mechanisms that shape the pathogen evolution and emergence, which is essential for development and implementation of intervention strategies.
Related collections
Most cited references29

- Record: found
- Abstract: found
- Article: found
IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies
- Record: found
- Abstract: found
- Article: not found
MUSCLE: multiple sequence alignment with high accuracy and high throughput.
- Record: found
- Abstract: found
- Article: not found
