- Record: found
- Abstract: found
- Article: found
JAK-STAT and G-protein-coupled receptor signaling pathways are frequently altered in epitheliotropic intestinal T-cell lymphoma
Read this article at
Abstract
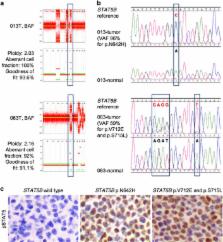
Epitheliotropic intestinal T-cell lymphoma (EITL, also known as type II enteropathy-associated T-cell lymphoma) is an aggressive intestinal disease with poor prognosis and its molecular alterations have not been comprehensively characterized. We aimed to identify actionable easy-to-screen alterations that would allow better diagnostics and/or treatment of this deadly disease. By performing whole-exome sequencing of four EITL tumor-normal pairs, followed by amplicon deep sequencing of 42 tumor samples, frequent alterations of the JAK-STAT and G-protein-coupled receptor (GPCR) signaling pathways were discovered in a large portion of samples. Specifically, STAT5B was mutated in a remarkable 63% of cases, JAK3 in 35% and GNAI2 in 24%, with the majority occurring at known activating hotspots in key functional domains. Moreover, STAT5B locus carried copy-neutral loss of heterozygosity resulting in the duplication of the mutant copy, suggesting the importance of mutant STAT5B dosage for the development of EITL. Dysregulation of the JAK-STAT and GPCR pathways was also supported by gene expression profiling and further verified in patient tumor samples. In vitro overexpression of GNAI2 mutants led to the upregulation of pERK1/2, a member of MEK-ERK pathway. Notably, inhibitors of both JAK-STAT and MEK-ERK pathways effectively reduced viability of patient-derived primary EITL cells, indicating potential therapeutic strategies for this neoplasm with no effective treatment currently available.
Related collections
Most cited references35
- Record: found
- Abstract: found
- Article: not found
Integration of gene mutations in risk prognostication for patients receiving first-line immunochemotherapy for follicular lymphoma: a retrospective analysis of a prospective clinical trial and validation in a population-based registry.
- Record: found
- Abstract: found
- Article: not found