- Record: found
- Abstract: found
- Article: not found
Modeling the underestimation of COVID-19 infection
Read this article at
Abstract
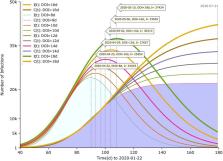
Estimation of the undocumented cases of COVID-19 is critical for understanding the epidemic potential of the disease and informing pandemic response. The COVID-19 pandemic originated from a severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a virus similar to severe acute respiratory syndrome (SARS) that was formerly identified in 2003. The contagiousness, dynamics of the pathogen, and mobility of the general population incurred the occurrence of underestimation of infection (i.e., the unidentified cases and the gap with the identified cases) that was potentially substantial in magnitude, which was supposed to connect with subsequent cyclical outbreaks in practice. We employed a Susceptible-Infected-Removed-Contained (SIR-C) mathematical model to infer critical epidemiological characteristics associated with COVID-19, then asymptotically simulated the peak sizes and peak dates of the identified and unidentified cases, the underestimation, and the dynamics of the gap. The simulation outcomes indicated that unidentified peak dates in practice could predate the reported peak dates for a variable period of weeks or months. In comparison, the saturation sizes of infection remained at commensurate levels. The curve of the initial exponential-like outbreak for the undocumented cases would flatten when the gap between concurrent identified cases and unidentified cases decreased. The rate of non-pharmaceutical containment could impact the trend of disease transmission ceteris paribus, and the greater the rate the larger reduction of infections. When the rate reached a certain level of threshold, the undocumented curve would shift from flattening effect to decaying effect. A similar trend was observed when it applied to the rate of pharmaceutical containment measures ceteris paribus. The results were sensitive to the duration of infection (DOI), it manifested that greater values of DOI were associated with greater peak sizes and greater peak dates for both documented and undocumented cases. Conditional on assumptions, calibration of DOI from 8 days to 18 days would increase the unidentified peak size by nearly 56% and the peak date by almost 18 days.
Related collections
Most cited references19
- Record: found
- Abstract: found
- Article: not found
Temporal dynamics in viral shedding and transmissibility of COVID-19
- Record: found
- Abstract: found
- Article: not found
Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study

- Record: found
- Abstract: found
- Article: found
