- Record: found
- Abstract: found
- Article: found
Deep learning enables automated MRI-based estimation of uterine volume also in patients with uterine fibroids undergoing high-intensity focused ultrasound therapy
Read this article at
Abstract
Background
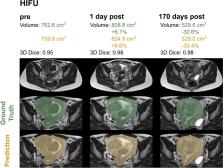
High-intensity focused ultrasound (HIFU) is used for the treatment of symptomatic leiomyomas. We aim to automate uterine volumetry for tracking changes after therapy with a 3D deep learning approach.
Methods
A 3D nnU-Net model in the default setting and in a modified version including convolutional block attention modules (CBAMs) was developed on 3D T2-weighted MRI scans. Uterine segmentation was performed in 44 patients with routine pelvic MRI (standard group) and 56 patients with uterine fibroids undergoing ultrasound-guided HIFU therapy (HIFU group). Here, preHIFU scans ( n = 56), postHIFU imaging maximum one day after HIFU ( n = 54), and the last available follow-up examination ( n = 53, days after HIFU: 420 ± 377) were included. The training was performed on 80% of the data with fivefold cross-validation. The remaining data were used as a hold-out test set. Ground truth was generated by a board-certified radiologist and a radiology resident. For the assessment of inter-reader agreement, all preHIFU examinations were segmented independently by both.
Results
High segmentation performance was already observed for the default 3D nnU-Net (mean Dice score = 0.95 ± 0.05) on the validation sets. Since the CBAM nnU-Net showed no significant benefit, the less complex default model was applied to the hold-out test set, which resulted in accurate uterus segmentation (Dice scores: standard group 0.92 ± 0.07; HIFU group 0.96 ± 0.02), which was comparable to the agreement between the two readers.
Key points
Related collections
Most cited references36
- Record: found
- Abstract: found
- Article: not found
3D Slicer as an image computing platform for the Quantitative Imaging Network.
- Record: found
- Abstract: found
- Article: found
seaborn: statistical data visualization
- Record: found
- Abstract: found
- Article: not found