- Record: found
- Abstract: found
- Article: found
Targeted next-generation sequencing for detection of PIK3CA mutations in archival tissues from patients with Klippel–Trenaunay syndrome in an Asian population : List the full names and institutional addresses for all authors
Read this article at
Abstract
Background
Klippel–Trenaunay syndrome (KTS) is a rare slow-flow combined vascular malformation with limb hypertrophy. KTS is thought to lie on the PIK3CA-related overgrowth spectrum, but reports are limited. PIK3CA encodes p110α, a catalytic subunit of phosphatidylinositol 3-kinase (PI3K) that plays an essential role in the PI3K/AKT/mammalian target of rapamycin (mTOR) signaling pathway. We aimed to demonstrate the clinical utility of targeted next-generation sequencing (NGS) in identifying PIK3CA mosaicism in archival formalin-fixed paraffin-embedded (FFPE) tissues from patients with KTS.
Results
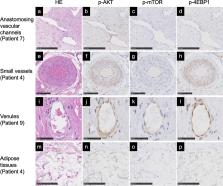
Participants were 9 female and 5 male patients with KTS diagnosed as capillaro-venous malformation (CVM) or capillaro-lymphatico-venous malformation (CLVM). Median age at resection was 14 years (range, 5–57 years). Median archival period before DNA extraction from FFPE tissues was 5.4 years (range, 3–7 years). NGS-based sequencing of PIK3CA achieved an amplicon mean coverage of 119,000x. PIK3CA missense mutations were found in 12 of 14 patients (85.7%; 6/8 CVM and 6/6 CLVM), with 8 patients showing the hotspot variants E542K, E545K, H1047R, and H1047L. The non-hotspot PIK3CA variants C420R, Q546K, and Q546R were identified in 4 patients. Overall, the mean variant allele frequency for identified PIK3CA variants was 6.9% (range, 1.6–17.4%). All patients with geographic capillary malformation, histopathological lymphatic malformation or macrodactyly of the foot had PIK3CA variants. No genotype–phenotype association between hotspot and non-hotspot PIK3CA variants was found. Histologically, the vessels and adipose tissues of the lesions showed phosphorylation of the proteins in the PI3K/AKT/mTOR signaling pathway, including p-AKT, p-mTOR, and p-4EBP1.
Conclusions
The PI3K/AKT/mTOR pathway in mesenchymal tissues was activated in patients with KTS. Amplicon-based targeted NGS could identify low-level mosaicism from low-input DNA extracted from FFPE tissues, potentially providing a diagnostic option for personalized medicine with inhibitors of the PI3K/AKT/mTOR signaling pathway.
Related collections
Most cited references66
- Record: found
- Abstract: found
- Article: found
High frequency of mutations of the PIK3CA gene in human cancers.
- Record: found
- Abstract: found
- Article: not found
Angiogenesis selectively requires the p110alpha isoform of PI3K to control endothelial cell migration.
- Record: found
- Abstract: found
- Article: not found