- Record: found
- Abstract: found
- Article: found
People of the British Isles: preliminary analysis of genotypes and surnames in a UK-control population
Abstract
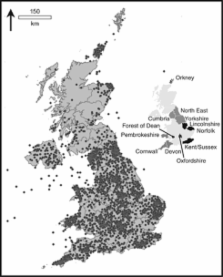
There is a great deal of interest in a fine-scale population structure in the UK, both as a signature of historical immigration events and because of the effect population structure may have on disease association studies. Although population structure appears to have a minor impact on the current generation of genome-wide association studies, it is likely to have a significant part in the next generation of studies designed to search for rare variants. A powerful way of detecting such structure is to control and document carefully the provenance of the samples involved. In this study, we describe the collection of a cohort of rural UK samples (The People of the British Isles), aimed at providing a well-characterised UK-control population that can be used as a resource by the research community, as well as providing a fine-scale genetic information on the British population. So far, some 4000 samples have been collected, the majority of which fit the criteria of coming from a rural area and having all four grandparents from approximately the same area. Analysis of the first 3865 samples that have been geocoded indicates that 75% have a mean distance between grandparental places of birth of 37.3 km, and that about 70% of grandparental places of birth can be classed as rural. Preliminary genotyping of 1057 samples demonstrates the value of these samples for investigating a fine-scale population structure within the UK, and shows how this can be enhanced by the use of surnames.
Related collections
Most cited references29
- Record: found
- Abstract: found
- Article: not found
A map of human genome variation from population-scale sequencing.
- Record: found
- Abstract: found
- Article: not found
Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls.
- Record: found
- Abstract: found
- Article: not found