- Record: found
- Abstract: found
- Article: found
Mutational Analysis of a Familial Adenomatous Polyposis Pedigree with Bile Duct Polyp Phenotype
Read this article at
Abstract
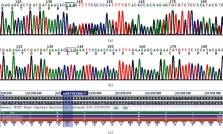
A large number of colorectal cancers have a genetic background in China. However, due to insufficient awareness, the diagnostic rate remains low and merely 5-6% of colorectal cancer patients are diagnosed with hereditary colorectal cancer. Familial adenomatous polyposis (FAP) is an autosomal dominant genetic disease caused by mutations in the adenomatous polyposis coli (APC) gene. Different mutation sites in APC are associated with the severity of FAP, risks of carcinogenesis, and extraintestinal manifestations. We used next-generation sequencing (NGS) and capture techniques to screen suspected mutation points in the proband in this pedigree. Using modified Sanger sequencing, we identified members of the family who were carriers of this variant and whether this segregated well with disease occurrence. FAP family members had multiple adenomatous polyps in their gastrointestinal tracts, some of which developed into cancer with age. Two subjects presented a rare common bile duct polyp phenotype. No extraintestinal manifestations were observed. A heterozygous frameshift mutation in APC exon 16 (NM_000038.6) was observed in the proband and in other patients: c.3260_3261del (p.Leu1087GlnQfs ∗ 31) (rs587782305); the variant call format was CCT/C. Due to the deletion of two bases, a stop codon appeared after 31 amino acids, and the protein was truncated prematurely, which affected the conformation of the protein. Pedigree genetic linkage analysis showed that the clinical phenotype cosegregated with the APC mutation p.L1087fs. This mutation may be the pathogenic in this FAP family and responsible for this rare common bile duct polyp.
Related collections
Most cited references44
- Record: found
- Abstract: found
- Article: not found
Correlations between mutation site in APC and phenotype of familial adenomatous polyposis (FAP): a review of the literature.
- Record: found
- Abstract: found
- Article: not found
Identification and characterization of the familial adenomatous polyposis coli gene
- Record: found
- Abstract: found
- Article: not found