- Record: found
- Abstract: found
- Article: not found
Peimine inhibits variants of SARS‐CoV‐2 cell entry via blocking the interaction between viral spike protein and ACE2

Read this article at
Abstract
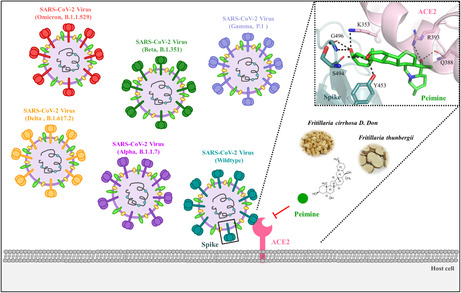
Coronavirus disease 2019 (COVID‐19) is caused by the novel severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2). Several vaccines against SARS‐CoV‐2 have been approved; however, variants of concern (VOCs) can evade vaccine protection. Therefore, developing small compound drugs that directly block the interaction between the viral spike glycoprotein and ACE2 is urgently needed to provide a complementary or alternative treatment for COVID‐19 patients. We developed a viral infection assay to screen a library of approximately 126 small molecules and showed that peimine inhibits VOCs viral infections. In addition, a fluorescence resonance energy transfer (FRET) assay showed that peimine suppresses the interaction of spike and ACE2. Molecular docking analysis revealed that peimine exhibits a higher binding affinity for variant spike proteins and is able to form hydrogen bonds with N501Y in the spike protein. These results suggest that peimine, a compound isolated from Fritillaria, may be a potent inhibitor of SARS‐CoV‐2 variant infection.
Practical applications
In this study, we identified a naturally derived compound of peimine, a major bioactive alkaloid extracted from Fritillaria, that could inhibit SARS‐CoV‐2 variants of concern (VOCs) viral infection in 293T/ACE2 and Calu‐3 lung cells. In addition, peimine blocks viral entry through interruption of spike and ACE2 interaction. Moreover, molecular docking analysis demonstrates that peimine has a higher binding affinity on N501Y in the spike protein. Furthermore, we found that Fritillaria significantly inhibits SARS‐CoV‐2 viral infection. These results suggested that peimine and Fritillaria could be a potential functional drug and food for COVID‐19 patients.
Abstract
Schematic model of the inhibitory mechanism of peimine for interactions between the viral spike protein and ACE2 on the cell surface. Peimine inhibits variants of concern (VOCs) of SARS‐CoV‐2 entry in 293T/ACE2 and Calu‐3 lung cells. Peimine blocks viral entry through interruption of spike and ACE2 interaction. Molecular docking analysis demonstrates that peimine has a higher binding affinity on N501Y in the spike protein.
Related collections
Most cited references57
- Record: found
- Abstract: found
- Article: not found
SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor
- Record: found
- Abstract: found
- Article: not found
Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor

- Record: found
- Abstract: found
- Article: found