- Record: found
- Abstract: found
- Article: found
Graph theory applications in congenital heart disease
Read this article at
Abstract
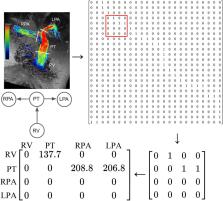
Graph theory can be used to address problems with complex network structures. Congenital heart diseases (CHDs) involve complex abnormal connections between chambers, vessels, and organs. We proposed a new method to represent CHDs based on graph theory, wherein vertices were defined as the spaces through which blood flows and edges were defined by the blood flow between the spaces and direction of the blood flow. The CHDs of tetralogy of Fallot (TOF) and transposition of the great arteries (TGA) were selected as examples for constructing directed graphs and binary adjacency matrices. Patients with totally repaired TOF, surgically corrected d-TGA, and Fontan circulation undergoing four-dimensional (4D) flow magnetic resonance imaging (MRI) were included as examples for constructing the weighted adjacency matrices. The directed graphs and binary adjacency matrices of the normal heart, extreme TOF undergoing a right modified Blalock–Taussig shunt, and d-TGA with a ventricular septal defect were constructed. The weighted adjacency matrix of totally repaired TOF was constructed using the peak velocities obtained from 4D flow MRI. The developed method is promising for representing CHDs and may be helpful in developing artificial intelligence and conducting future research on CHD.
Related collections
Most cited references37
- Record: found
- Abstract: not found
- Article: not found
Complex brain networks: graph theoretical analysis of structural and functional systems.
- Record: found
- Abstract: found
- Article: not found
Neurophysiological architecture of functional magnetic resonance images of human brain.

