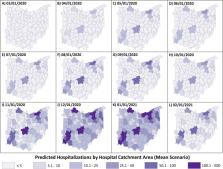
Summary Background In the face of rapidly changing data, a range of case fatality ratio estimates for coronavirus disease 2019 (COVID-19) have been produced that differ substantially in magnitude. We aimed to provide robust estimates, accounting for censoring and ascertainment biases. Methods We collected individual-case data for patients who died from COVID-19 in Hubei, mainland China (reported by national and provincial health commissions to Feb 8, 2020), and for cases outside of mainland China (from government or ministry of health websites and media reports for 37 countries, as well as Hong Kong and Macau, until Feb 25, 2020). These individual-case data were used to estimate the time between onset of symptoms and outcome (death or discharge from hospital). We next obtained age-stratified estimates of the case fatality ratio by relating the aggregate distribution of cases to the observed cumulative deaths in China, assuming a constant attack rate by age and adjusting for demography and age-based and location-based under-ascertainment. We also estimated the case fatality ratio from individual line-list data on 1334 cases identified outside of mainland China. Using data on the prevalence of PCR-confirmed cases in international residents repatriated from China, we obtained age-stratified estimates of the infection fatality ratio. Furthermore, data on age-stratified severity in a subset of 3665 cases from China were used to estimate the proportion of infected individuals who are likely to require hospitalisation. Findings Using data on 24 deaths that occurred in mainland China and 165 recoveries outside of China, we estimated the mean duration from onset of symptoms to death to be 17·8 days (95% credible interval [CrI] 16·9–19·2) and to hospital discharge to be 24·7 days (22·9–28·1). In all laboratory confirmed and clinically diagnosed cases from mainland China (n=70 117), we estimated a crude case fatality ratio (adjusted for censoring) of 3·67% (95% CrI 3·56–3·80). However, after further adjusting for demography and under-ascertainment, we obtained a best estimate of the case fatality ratio in China of 1·38% (1·23–1·53), with substantially higher ratios in older age groups (0·32% [0·27–0·38] in those aged <60 years vs 6·4% [5·7–7·2] in those aged ≥60 years), up to 13·4% (11·2–15·9) in those aged 80 years or older. Estimates of case fatality ratio from international cases stratified by age were consistent with those from China (parametric estimate 1·4% [0·4–3·5] in those aged <60 years [n=360] and 4·5% [1·8–11·1] in those aged ≥60 years [n=151]). Our estimated overall infection fatality ratio for China was 0·66% (0·39–1·33), with an increasing profile with age. Similarly, estimates of the proportion of infected individuals likely to be hospitalised increased with age up to a maximum of 18·4% (11·0–7·6) in those aged 80 years or older. Interpretation These early estimates give an indication of the fatality ratio across the spectrum of COVID-19 disease and show a strong age gradient in risk of death. Funding UK Medical Research Council.