- Record: found
- Abstract: found
- Article: found
Single-Nucleotide Polymorphism Genotyping Identifies a Locally Endemic Clone of Methicillin-Resistant Staphylococcus aureus
Read this article at
Abstract
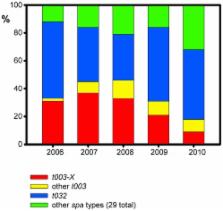
We developed, tested, and applied a TaqMan real-time PCR assay for interrogation of three single-nucleotide polymorphisms that differentiate a clade (termed ‘ t003-X’) within the radiation of methicillin-resistant Staphylococcus aureus (MRSA) ST225. The TaqMan assay achieved 98% typeability and results were fully concordant with DNA sequencing. By applying this assay to 305 ST225 isolates from an international collection, we demonstrate that clade t003-X is endemic in a single acute-care hospital in Germany at least since 2006, where it has caused a substantial proportion of infections. The strain was also detected in another hospital located 16 kilometers away. Strikingly, however, clade t003-X was not found in 62 other hospitals throughout Germany nor among isolates from other countries, and, hence, displayed a very restricted geographical distribution. Consequently, our results show that SNP-typing may be useful to identify and track MRSA clones that are specific to individual healthcare institutions. In contrast, the spatial dissemination pattern observed here had not been resolved by other typing procedures, including multilocus sequence typing (MLST), spa typing, DNA macrorestriction, and multilocus variable-number tandem repeat analysis (MLVA).
Related collections
Most cited references28
- Record: found
- Abstract: found
- Article: not found
The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA).
- Record: found
- Abstract: found
- Article: not found
Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database.
- Record: found
- Abstract: found
- Article: not found