- Record: found
- Abstract: found
- Article: not found
Ultrastructural modifications induced by SARS-CoV-2 in Vero cells: a kinetic analysis of viral factory formation, viral particle morphogenesis and virion release
Read this article at
Abstract
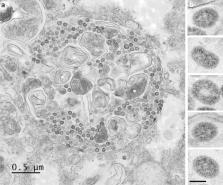
Many studies on SARS-CoV-2 have been performed over short-time scale, but few have focused on the ultrastructural characteristics of infected cells. We used TEM to perform kinetic analysis of the ultrastructure of SARS-CoV-2-infected cells. Early infection events were characterized by the presence of clusters of single-membrane vesicles and stacks of membrane containing nuclear pores called annulate lamellae (AL). A large network of host cell-derived organelles transformed into virus factories was subsequently observed in the cells. As previously described for other RNA viruses, these replication factories consisted of double-membrane vesicles (DMVs) located close to the nucleus. Viruses released at the cell surface by exocytosis harbored the typical crown of spike proteins, but viral particles without spikes were also observed in intracellular compartments, possibly reflecting incorrect assembly or a cell degradation process.
Related collections
Most cited references38
- Record: found
- Abstract: found
- Article: not found
A Novel Coronavirus from Patients with Pneumonia in China, 2019

- Record: found
- Abstract: found
- Article: found
A pneumonia outbreak associated with a new coronavirus of probable bat origin
- Record: found
- Abstract: found
- Article: not found
