- Record: found
- Abstract: found
- Article: found
Combined analyses of kinship and F ST suggest potential drivers of chaotic genetic patchiness in high gene-flow populations
Read this article at
Abstract
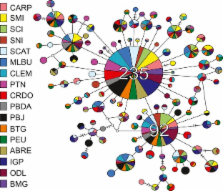
We combine kinship estimates with traditional F-statistics to explain contemporary drivers of population genetic differentiation despite high gene flow. We investigate range-wide population genetic structure of the California spiny (or red rock) lobster ( Panulirus interruptus) and find slight, but significant global population differentiation in mtDNA (Φ ST = 0.006, P = 0.001; D est_Chao = 0.025) and seven nuclear microsatellites ( F ST = 0.004, P < 0.001; D est_Chao = 0.03), despite the species’ 240- to 330-day pelagic larval duration. Significant population structure does not correlate with distance between sampling locations, and pairwise F ST between adjacent sites often exceeds that among geographically distant locations. This result would typically be interpreted as unexplainable, chaotic genetic patchiness. However, kinship levels differ significantly among sites (pseudo- F 16,988 = 1.39, P = 0.001), and ten of 17 sample sites have significantly greater numbers of kin than expected by chance ( P < 0.05). Moreover, a higher proportion of kin within sites strongly correlates with greater genetic differentiation among sites ( D est_Chao, R 2 = 0.66, P < 0.005). Sites with elevated mean kinship were geographically proximate to regions of high upwelling intensity ( R 2 = 0.41, P = 0.0009). These results indicate that P. interruptus does not maintain a single homogenous population, despite extreme dispersal potential. Instead, these lobsters appear to either have substantial localized recruitment or maintain planktonic larval cohesiveness whereby siblings more likely settle together than disperse across sites. More broadly, our results contribute to a growing number of studies showing that low F ST and high family structure across populations can coexist, illuminating the foundations of cryptic genetic patterns and the nature of marine dispersal.
Related collections
Most cited references190
- Record: found
- Abstract: found
- Article: not found
DnaSP, DNA polymorphism analyses by the coalescent and other methods.
- Record: found
- Abstract: found
- Article: not found
What is a population? An empirical evaluation of some genetic methods for identifying the number of gene pools and their degree of connectivity.
- Record: found
- Abstract: found
- Article: not found