- Record: found
- Abstract: found
- Article: found
T-cell metagene predicts a favorable prognosis in estrogen receptor-negative and HER2-positive breast cancers
Read this article at
Abstract
Introduction
Lymphocyte infiltration (LI) is often seen in breast cancer but its importance remains controversial. A positive correlation of human epidermal growth factor receptor 2 (HER2) amplification and LI has been described, which was associated with a more favorable outcome. However, specific lymphocytes might also promote tumor progression by shifting the cytokine milieu in the tumor.
Methods
Affymetrix HG-U133A microarray data of 1,781 primary breast cancer samples from 12 datasets were included. The correlation of immune system-related metagenes with different immune cells, clinical parameters, and survival was analyzed.
Results
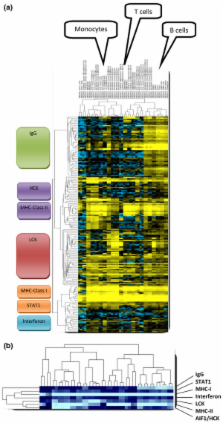
A large cluster of nearly 600 genes with functions in immune cells was consistently obtained in all datasets. Seven robust metagenes from this cluster can act as surrogate markers for the amount of different immune cell types in the breast cancer sample. An IgG metagene as a marker for B cells had no significant prognostic value. In contrast, a strong positive prognostic value for the T-cell surrogate marker (lymphocyte-specific kinase (LCK) metagene) was observed among all estrogen receptor (ER)-negative tumors and those ER-positive tumors with a HER2 overexpression. Moreover ER-negative tumors with high expression of both IgG and LCK metagenes seem to respond better to neoadjuvant chemotherapy.
Conclusions
Precise definitions of the specific subtypes of immune cells in the tumor can be accomplished from microarray data. These surrogate markers define subgroups of tumors with different prognosis. Importantly, all known prognostic gene signatures uniformly assign poor prognosis to all ER-negative tumors. In contrast, the LCK metagene actually separates the ER-negative group into better or worse prognosis.
Related collections
Most cited references42
- Record: found
- Abstract: found
- Article: found
Stromal gene expression predicts clinical outcome in breast cancer.
- Record: found
- Abstract: found
- Article: not found
Strong Time Dependence of the 76-Gene Prognostic Signature for Node-Negative Breast Cancer Patients in the TRANSBIG Multicenter Independent Validation Series
- Record: found
- Abstract: found
- Article: not found