- Record: found
- Abstract: found
- Article: found
MiR-21 regulating PVT1/PTEN/IL-17 axis towards the treatment of infectious diabetic wound healing by modified GO-derived biomaterial in mouse models
Read this article at
Abstract
Background
Diabetic foot ulcer (DFU), persistent hyperglycemia and inflammation, together with impaired nutrient and oxygen deficiency, can present abnormal angiogenesis following tissue injury such that these tissues fail to heal properly. It is critical to design a new treatment method for DFU patients with a distinct biomechanism that is more effective than current treatment regimens.
Method
Graphene oxide (GO) was combined with a biocompatible polymer as a kind of modified GO-based hydrogel. The characterization of our biomaterial was measured in vitro. The repair efficiency of the biomaterial was evaluated in the mouse full-skin defect models. The key axis related to diabetic wound (DW) was identified and investigated using bioinformatics analyses and practical experiments.
Result
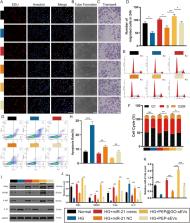
In the study, we found that our modified GO-based wound dressing material is a promising option for diabetic wound. Secondly, our biomaterial could enhance the secretion of small EVs (sEVs) with more miR-21 by adipose-derived mesenchymal stem cells (AD-MSCs). Thirdly, the PVT1/PTEN/IL-17 axis was found to be decreased to promote DFU wound healing by modifying miR-21 with the discovery of PVT1 as a critical LncRNA by bioinformatics analysis and tests.
Related collections
Most cited references58
- Record: found
- Abstract: found
- Article: not found
The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses.
- Record: found
- Abstract: found
- Article: not found
Exosomes Facilitate Therapeutic Targeting of Oncogenic Kras in Pancreatic Cancer

- Record: found
- Abstract: found
- Article: found