- Record: found
- Abstract: found
- Article: found
Performance and impact of a multiplex PCR in ICU patients with ventilator-associated pneumonia or ventilated hospital-acquired pneumonia
Read this article at
Abstract
Background
Early appropriate antibiotic therapy reduces morbidity and mortality of severe pneumonia. However, the emergence of bacterial resistance requires the earliest use of antibiotics with the narrowest possible spectrum. The Unyvero Hospitalized Pneumonia (HPN, Curetis) test is a multiplex PCR (M-PCR) system detecting 21 bacteria and 19 resistance genes on respiratory samples within 5 h. We assessed the performance and the potential impact of the M-PCR on the antibiotic therapy of ICU patients.
Methods
In this prospective study, we performed a M-PCR on bronchoalveolar lavage (BAL) or plugged telescoping catheter (PTC) samples of patients with ventilated HAP or VAP with Gram-negative bacilli or clustered Gram-positive cocci. This study was conducted in 3 ICUs in a French academic hospital: the medical and infectious diseases ICU, the surgical ICU, and the cardio-surgical ICU. A multidisciplinary expert panel simulated the antibiotic changes they would have made if the M-PCR results had been available.
Results
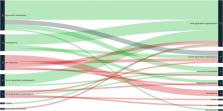
We analyzed 95 clinical samples of ventilated HAP or VAP (72 BAL and 23 PTC) from 85 patients (62 males, median age 64 years). The median turnaround time of the M-PCR was 4.6 h (IQR 4.4–5). A total of 90/112 bacteria were detected by the M-PCR system with a global sensitivity of 80% (95% CI, 73–88%) and specificity of 99% (95% CI 99–100). The sensitivity was better for Gram-negative bacteria (90%) than for Gram-positive cocci (62%) ( p = 0.005). Moreover, 5/8 extended-spectrum beta-lactamases (CTX-M gene) and 4/4 carbapenemases genes (3 NDM, one oxa-48) were detected. The M-PCR could have led to the earlier initiation of an effective antibiotic in 20/95 patients (21%) and to early de-escalation in 37 patients (39%) but could also have led to one (1%) inadequate antimicrobial therapy. Among 17 empiric antibiotic treatments with carbapenems, 10 could have been de-escalated in the following hours according to the M-PCR results. The M-PCR also led to 2 unexpected diagnosis of severe legionellosis confirmed by culture methods.
Related collections
Most cited references25
- Record: found
- Abstract: found
- Article: not found
Changes in Prevalence of Health Care–Associated Infections in U.S. Hospitals
- Record: found
- Abstract: found
- Article: not found
Randomized Trial of Rapid Multiplex Polymerase Chain Reaction-Based Blood Culture Identification and Susceptibility Testing.

- Record: found
- Abstract: found
- Article: found