- Record: found
- Abstract: found
- Article: found
A comprehensive analysis of the faecal microbiome and metabolome of Strongyloides stercoralis infected volunteers from a non-endemic area
Read this article at
Abstract
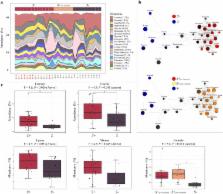
Data from recent studies support the hypothesis that infections by human gastrointestinal (GI) helminths impact, directly and/or indirectly, on the composition of the host gut microbial flora. However, to the best of our knowledge, these studies have been conducted in helminth-endemic areas with multi-helminth infections and/or in volunteers with underlying gut disorders. Therefore, in this study, we explore the impact of natural mono-infections by the human parasite Strongyloides stercoralis on the faecal microbiota and metabolic profiles of a cohort of human volunteers from a non-endemic area of northern Italy ( S+), pre- and post-anthelmintic treatment, and compare the findings with data obtained from a cohort of uninfected controls from the same geographical area ( S−). Analyses of bacterial 16S rRNA high-throughput sequencing data revealed increased microbial alpha diversity and decreased beta diversity in the faecal microbial profiles of S+ subjects compared to S−. Furthermore, significant differences in the abundance of several bacterial taxa were observed between samples from S+ and S− subjects, and between S+ samples collected pre- and post-anthelmintic treatment. Faecal metabolite analysis detected marked increases in the abundance of selected amino acids in S+ subjects, and of short chain fatty acids in S− subjects. Overall, our work adds valuable knowledge to current understanding of parasite-microbiota associations and will assist future mechanistic studies aimed to unravel the causality of these relationships.
Related collections
Most cited references66

- Record: found
- Abstract: found
- Article: found
Through Ageing, and Beyond: Gut Microbiota and Inflammatory Status in Seniors and Centenarians

- Record: found
- Abstract: found
- Article: found
Archaea and Fungi of the Human Gut Microbiome: Correlations with Diet and Bacterial Residents
- Record: found
- Abstract: found
- Article: found