- Record: found
- Abstract: found
- Article: found
Complete Sequencing of pNDM-HK Encoding NDM-1 Carbapenemase from a Multidrug-Resistant Escherichia coli Strain Isolated in Hong Kong
Read this article at
Abstract
Background
The emergence of plasmid-mediated carbapenemases, such as NDM-1 in Enterobacteriaceae is a major public health issue. Since they mediate resistance to virtually all β-lactam antibiotics and there is often co-resistance to other antibiotic classes, the therapeutic options for infections caused by these organisms are very limited.
Methodology
We characterized the first NDM-1 producing E. coli isolate recovered in Hong Kong. The plasmid encoding the metallo-β-lactamase gene was sequenced.
Principal Findings
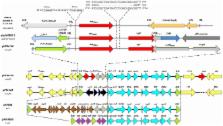
The plasmid, pNDM-HK readily transferred to E. coli J53 at high frequencies. It belongs to the broad host range IncL/M incompatibility group and is 88803 bp in size. Sequence alignment showed that pNDM-HK has a 55 kb backbone which shared 97% homology with pEL60 originating from the plant pathogen, Erwina amylovora in Lebanon and a 28.9 kb variable region. The plasmid backbone includes the mucAB genes mediating ultraviolet light resistance. The 28.9 kb region has a composite transposon-like structure which includes intact or truncated genes associated with resistance to β-lactams ( bla TEM-1, bla NDM-1, Δ bla DHA-1), aminoglycosides ( aacC2, armA), sulphonamides ( sul1) and macrolides ( mel, mph2). It also harbors the following mobile elements: IS 26, IS CR1, tnpU, tnpAcp2, tnpD, Δ tnpATn 1 and insL. Certain blocks within the 28.9 kb variable region had homology with the corresponding sequences in the widely disseminated plasmids, pCTX-M3, pMUR050 and pKP048 originating from bacteria in Poland in 1996, in Spain in 2002 and in China in 2006, respectively.
Related collections
Most cited references29
- Record: found
- Abstract: found
- Article: not found
Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study
- Record: found
- Abstract: found
- Article: not found
ISCR elements: novel gene-capturing systems of the 21st century?
- Record: found
- Abstract: found
- Article: not found