- Record: found
- Abstract: found
- Article: not found
In-Solution IgG Titer Determination in Fermentation Broth Using Affibodies and Flow-Induced Dispersion Analysis
Read this article at
Abstract
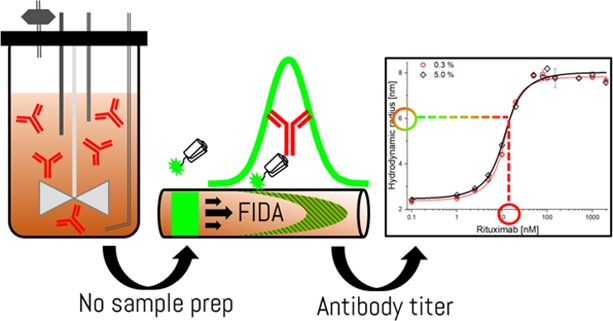
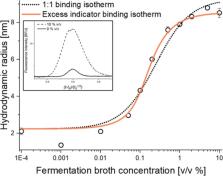
Biopharmaceuticals such as protein and peptide-based drugs are often produced by fermentation processes where it is necessary to monitor the amount and quality of the product expressed during fermentation and for release testing of the final drug product. Standard procedures involve surface-based ligand binding technologies such as enzyme-linked immunosorbent assay and biolayer interferometry, or extensive purification using, e.g., preparative chromatography followed by spectrophotometric protein quantification. The multistep nature of these methodologies leads to lengthy protocols and renders real-time process control impractical. Recently, flow-induced dispersion analysis (FIDA) was introduced as a novel in-solution ligand binding technology, requiring only nano/microliter sample volumes. FIDA is based on Taylor dispersion analysis in narrow fused silica capillaries and provides the hydrodynamic radius of the binding ligand and complex in addition to the detailed binding characterization. Here, we demonstrate the use of FIDA for quantification of monoclonal IgG antibodies (rituximab) directly in mammalian cell fermentation broth with only 4 min of analysis time. The FIDA assay utilizes a small anti-IgG affibody, conjugated to a fluorophore, as a selective rituximab binder. The apparent change in the hydrodynamic radius of the affibody, as it interacts with known concentrations of rituximab, is used for generating a binding curve in a blank fermentation medium, and hence determining the dissociation constant and complex size. Finally, the binding curve is utilized for quantifying the rituximab titer concentration in clarified fermentation broth samples.
Related collections
Most cited references19
- Record: found
- Abstract: not found
- Article: not found
Opportunities and challenges of real-time release testing in biopharmaceutical manufacturing
- Record: found
- Abstract: found
- Article: not found
High-throughput work flow for IgG Fc-glycosylation analysis of biotechnological samples.
- Record: found
- Abstract: found
- Article: not found
