- Record: found
- Abstract: found
- Article: found
Paradigm shift in the diagnosis of peste des petits ruminants: scoping review
Read this article at
Abstract
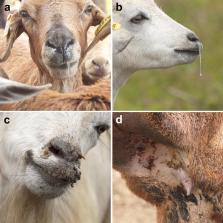
Peste des petits ruminants virus causes a highly contagious disease, which poses enormous economic losses in domestic animals and threatens the conservation of wild herbivores. Diagnosis remains a cornerstone to the Peste des petits ruminants Global Control and Eradication Strategy, an initiative of the World Organisation for Animal Health and the Food and Agriculture Organisation. The present review presents the peste des petits ruminants diagnostic landscape, including the practicality of commercially available diagnostic tools, prototype tests and opportunities for new technologies. The most common peste des petits ruminants diagnostic tools include; agar gel immunodiffusion, counter-immunoelectrophoresis, enzyme-linked immunosorbent assays, reverse transcription polymerase chain reaction either gel-based or real-time, reverse transcription loop-mediated isothermal amplification, reverse transcription recombinase polymerase amplification assays, immunochromatographic lateral flow devices, luciferase immunoprecipitation system and pseudotype-based assays. These tests vary in their technical demands, but all require a laboratory with exception of immunochromatographic lateral flow and possibly reverse transcription loop-mediated isothermal amplification and reverse transcription recombinase polymerase amplification assays. Thus, we are proposing an efficient integration of diagnostic tests for rapid and correct identification of peste des petits ruminants in endemic zones and to rapidly confirm outbreaks. Deployment of pen-side tests will improve diagnostic capacity in extremely remote settings and susceptible wildlife ecosystems, where transportation of clinical samples in the optimum cold chain is unreliable.
Related collections
Most cited references116
- Record: found
- Abstract: found
- Article: not found
Surface Plasmon Resonance Clinical Biosensors for Medical Diagnostics.
- Record: found
- Abstract: found
- Article: not found
A first look at the Oxford Nanopore MinION sequencer.
- Record: found
- Abstract: found
- Article: not found