- Record: found
- Abstract: found
- Article: not found
Identification of a Novel Risk Locus for Multiple Sclerosis at 13q31.3 by a Pooled Genome-Wide Scan of 500,000 Single Nucleotide Polymorphisms
Read this article at
Abstract
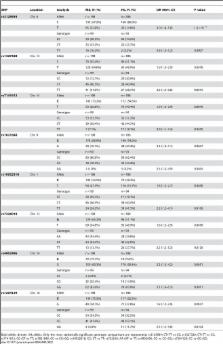
Multiple sclerosis is a chronic inflammatory demyelinating disease of the central nervous system with an important genetic component and strongest association driven by the HLA genes. We performed a pooling-based genome-wide association study of 500,000 SNPs in order to find new loci associated with the disease. After applying several criteria, 320 SNPs were selected from the microarrays and individually genotyped in a first and independent Spanish Caucasian replication cohort. The 8 most significant SNPs validated in this cohort were also genotyped in a second US Caucasian replication cohort for confirmation. The most significant association was obtained for SNP rs3129934, which neighbors the HLA-DRB/DQA loci and validates our pooling-based strategy. The second strongest association signal was found for SNP rs1327328, which resides in an unannotated region of chromosome 13 but is in linkage disequilibrium with nearby functional elements that may play important roles in disease susceptibility. This region of chromosome 13 has not been previously identified in MS linkage genome screens and represents a novel risk locus for the disease.
Related collections
Most cited references19
- Record: found
- Abstract: found
- Article: not found
Risk alleles for multiple sclerosis identified by a genomewide study.
- Record: found
- Abstract: found
- Article: not found
A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC.
- Record: found
- Abstract: found
- Article: not found
