- Record: found
- Abstract: found
- Article: found
Sequential Fragmentation of Pleistocene Forests in an East Africa Biodiversity Hotspot: Chameleons as a Model to Track Forest History
Read this article at
Abstract
Background
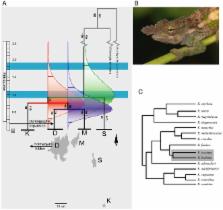
The Eastern Arc Mountains (EAM) is an example of naturally fragmented tropical forests, which contain one of the highest known concentrations of endemic plants and vertebrates. Numerous paleo-climatic studies have not provided direct evidence for ancient presence of Pleistocene forests, particularly in the regions in which savannah presently occurs. Knowledge of the last period when forests connected EAM would provide a sound basis for hypothesis testing of vicariance and dispersal models of speciation. Dated phylogenies have revealed complex patterns throughout EAM, so we investigated divergence times of forest fauna on four montane isolates in close proximity to determine whether forest break-up was most likely to have been simultaneous or sequential, using population genetics of a forest restricted arboreal chameleon, Kinyongia boehmei.
Methodology/Principal Findings
We used mitochondrial and nuclear genetic sequence data and mutation rates from a fossil-calibrated phylogeny to estimate divergence times between montane isolates using a coalescent approach. We found that chameleons on all mountains are most likely to have diverged sequentially within the Pleistocene from 0.93–0.59 Ma (95% HPD 0.22–1.84 Ma). In addition, post-hoc tests on chameleons on the largest montane isolate suggest a population expansion ∼182 Ka.
Conclusions/Significance
Sequential divergence is most likely to have occurred after the last of three wet periods within the arid Plio-Pleistocene era, but was not correlated with inter-montane distance. We speculate that forest connection persisted due to riparian corridors regardless of proximity, highlighting their importance in the region's historic dispersal events. The population expansion coincides with nearby volcanic activity, which may also explain the relative paucity of the Taita's endemic fauna. Our study shows that forest chameleons are an apposite group to track forest fragmentation, with the inference that forest extended between some EAM during the Pleistocene 1.1–0.9 Ma.
Related collections
Most cited references111
- Record: found
- Abstract: found
- Article: not found
MRBAYES: Bayesian inference of phylogenetic trees.
- Record: found
- Abstract: not found
- Article: not found
Speciation in amazonian forest birds.
- Record: found
- Abstract: found
- Article: not found