- Record: found
- Abstract: found
- Article: found
Using deep learning to associate human genes with age-related diseases
Read this article at
Abstract
Motivation
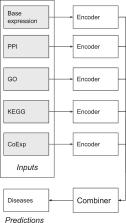
One way to identify genes possibly associated with ageing is to build a classification model (from the machine learning field) capable of classifying genes as associated with multiple age-related diseases. To build this model, we use a pre-compiled list of human genes associated with age-related diseases and apply a novel Deep Neural Network (DNN) method to find associations between gene descriptors (e.g. Gene Ontology terms, protein–protein interaction data and biological pathway information) and age-related diseases.
Results
The novelty of our new DNN method is its modular architecture, which has the capability of combining several sources of biological data to predict which ageing-related diseases a gene is associated with (if any). Our DNN method achieves better predictive performance than standard DNN approaches, a Gradient Boosted Tree classifier (a strong baseline method) and a Logistic Regression classifier. Given the DNN model produced by our method, we use two approaches to identify human genes that are not known to be associated with age-related diseases according to our dataset. First, we investigate genes that are close to other disease-associated genes in a complex multi-dimensional feature space learned by the DNN algorithm. Second, using the class label probabilities output by our DNN approach, we identify genes with a high probability of being associated with age-related diseases according to the model. We provide evidence of these putative associations retrieved from the DNN model with literature support.
Availability and implementation
The source code and datasets can be found at: https://github.com/fabiofabris/Bioinfo2019.
Related collections
Most cited references21
- Record: found
- Abstract: not found
- Article: not found
The genetic association database.
- Record: found
- Abstract: found
- Article: not found