- Record: found
- Abstract: found
- Article: found
A rare Waxy allele coordinately improves rice eating and cooking quality and grain transparency
Read this article at
Abstract
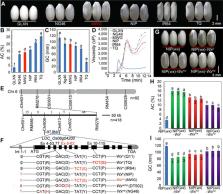
In rice ( Oryza sativa), amylose content (AC) is the major factor that determines eating and cooking quality (ECQ). The diversity in AC is largely attributed to natural allelic variation at the Waxy ( Wx) locus. Here we identified a rare Wx allele, Wx mw , which combines a favorable AC, improved ECQ and grain transparency. Based on a phylogenetic analysis of Wx genomic sequences from 370 rice accessions, we speculated that Wx mw may have derived from recombination between two important natural Wx alleles, Wx in and Wx b . We validated the effects of Wx mw on rice grain quality using both transgenic lines and near‐isogenic lines (NILs). When introgressed into the japonica Nipponbare (NIP) background, Wx mw resulted in a moderate AC that was intermediate between that of NILs carrying the Wx b allele and NILs with the Wx mp allele. Notably, mature grains of NILs fixed for Wx mw had an improved transparent endosperm relative to soft rice. Further, we introduced Wx mw into a high‐yielding japonica cultivar via molecular marker‐assisted selection: the introgressed lines exhibited clear improvements in ECQ and endosperm transparency. Our results suggest that Wx mw is a promising allele to improve grain quality, especially ECQ and grain transparency of high‐yielding japonica cultivars, in rice breeding programs.
Abstract
In the present study, a rare Wx allele, Wx mw which may have derived from recombination between two important natural Wx alleles in rice, was identified as a promising allele to improve grain quality, especially cooked rice taste and grain transparency of high‐yielding japonica cultivars, in rice breeding programs.
Related collections
Most cited references53
- Record: found
- Abstract: found
- Article: not found
Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method.
- Record: found
- Abstract: found
- Article: not found
MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms.
- Record: found
- Abstract: found
- Article: not found
