- Record: found
- Abstract: found
- Article: found
Structural investigation of a chaperonin in action reveals how nucleotide binding regulates the functional cycle
Read this article at
Abstract
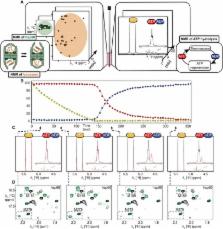
Site-selective isotope labeling enables structural and functional investigation of a working 1-MDa chaperonin by NMR spectroscopy.
Abstract
Chaperonins are ubiquitous protein assemblies present in bacteria, eukaryota, and archaea, facilitating the folding of proteins, preventing protein aggregation, and thus participating in maintaining protein homeostasis in the cell. During their functional cycle, they bind unfolded client proteins inside their double ring structure and promote protein folding by closing the ring chamber in an adenosine 5′-triphosphate (ATP)–dependent manner. Although the static structures of fully open and closed forms of chaperonins were solved by x-ray crystallography or electron microscopy, elucidating the mechanisms of such ATP-driven molecular events requires studying the proteins at the structural level under working conditions. We introduce an approach that combines site-specific nuclear magnetic resonance observation of very large proteins, enabled by advanced isotope labeling methods, with an in situ ATP regeneration system. Using this method, we provide functional insight into the 1-MDa large hsp60 chaperonin while processing client proteins and reveal how nucleotide binding, hydrolysis, and release control switching between closed and open states. While the open conformation stabilizes the unfolded state of client proteins, the internalization of the client protein inside the chaperonin cavity speeds up its functional cycle. This approach opens new perspectives to study structures and mechanisms of various ATP-driven biological machineries in the heat of action.
Related collections
Most cited references45
- Record: found
- Abstract: found
- Article: not found
Heat Shock Proteins and Cancer.
- Record: found
- Abstract: found
- Article: not found
Cross-correlated relaxation enhanced 1H[bond]13C NMR spectroscopy of methyl groups in very high molecular weight proteins and protein complexes.
- Record: found
- Abstract: found
- Article: not found