- Record: found
- Abstract: found
- Article: found
The Anti-Inflammatory Effects of Flavanol-Rich Lychee Fruit Extract in Rat Hepatocytes
Read this article at
Abstract
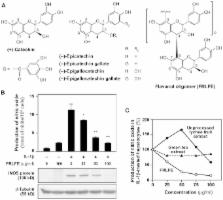
Flavanol (flavan-3-ol)-rich lychee fruit extract (FRLFE) is a mixture of oligomerized polyphenols primarily derived from lychee fruit and is rich in flavanol monomers, dimers, and trimers. Supplementation with this functional food has been shown to suppress inflammation and tissue damage caused by high-intensity exercise training. However, it is unclear whether FRLFE has in vitro anti-inflammatory effects, such as suppressing the production of the proinflammatory cytokine tumor necrosis factor α (TNF-α) and the proinflammatory mediator nitric oxide (NO), which is synthesized by inducible nitric oxide synthase (iNOS). Here, we analyzed the effects of FRLFE and its constituents on the expression of inflammatory genes in interleukin 1β (IL-1β)-treated rat hepatocytes. FRLFE decreased the mRNA and protein expression of the iNOS gene, leading to the suppression of IL-1β-induced NO production. FRLFE also decreased the levels of the iNOS antisense transcript, which stabilizes iNOS mRNA. By contrast, unprocessed lychee fruit extract, which is rich in flavanol polymers, and flavanol monomers had little effect on NO production. When a construct harboring the iNOS promoter fused to the firefly luciferase gene was used, FRLFE decreased the luciferase activity in the presence of IL-1β, suggesting that FRLFE suppresses the promoter activity of the iNOS gene at the transcriptional level. Electrophoretic mobility shift assays indicated that FRLFE reduced the nuclear transport of a key regulator, nuclear factor κB (NF-κB). Furthermore, FRLFE inhibited the phosphorylation of NF-κB inhibitor α (IκB-α). FRLFE also reduced the mRNA levels of NF-κB target genes encoding cytokines and chemokines, such as TNF-α. Therefore, FRLFE inhibited NF-κB activation and nuclear translocation to suppress the expression of these inflammatory genes. Our results suggest that flavanols may be responsible for the anti-inflammatory and hepatoprotective effects of FRLFE and may be used to treat inflammatory diseases.
Related collections
Most cited references37
- Record: found
- Abstract: not found
- Article: not found
Analysis of nitrate, nitrite, and [15N]nitrate in biological fluids.
- Record: found
- Abstract: found
- Article: not found
Analysis of microarray data using Z score transformation.
- Record: found
- Abstract: found
- Article: not found