- Record: found
- Abstract: found
- Article: not found
Intronic polyadenylation of PDGFRα in resident stem cells attenuates muscle fibrosis
Abstract
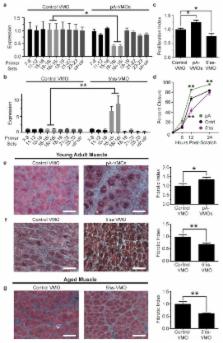
The platelet-derived growth factor receptor alpha (PDGFRα) exhibits divergent effects in skeletal muscle. At physiological levels, signaling through this receptor promotes muscle development in growing embryos and proper angiogenesis in regenerating adult muscle. 1, 2 However, both increased PDGF ligand abundance and enhanced PDGFRα pathway activity cause pathological fibrosis. 3, 4 This excessive collagen deposition, which is seen in aged and diseased muscle, 5– 7 interferes with proper muscle function and limits the effectiveness of gene- and cell-based therapies for muscle disorders. 8, 9 Although compelling evidence exists for the role of PDGFRα in fibrosis, little is known about the cells through which this pathway acts. Here we show that PDGFRα signaling regulates a population of muscle-resident fibro/adipogenic progenitors (FAPs) that play a supportive role in muscle regeneration but may also cause fibrosis when aberrantly regulated. 10– 13 We found that FAPs produce multiple transcriptional variants of PDGFRα with different polyadenylation sites, including an intronic variant that codes for a protein isoform containing a truncated kinase domain. This variant, upregulated during regeneration, acts as a decoy to inhibit PDGF signaling and to prevent FAP over-activation. Moreover, increasing expression of this isoform limits fibrosis in vivo, suggesting both biological relevance and therapeutic potential of modulating polyadenylation patterns in stem cell populations.
Related collections
Most cited references18
- Record: found
- Abstract: found
- Article: not found
PDGFRA activating mutations in gastrointestinal stromal tumors.
- Record: found
- Abstract: found
- Article: not found
Direct RNA sequencing.
- Record: found
- Abstract: found
- Article: not found
